 This is my fourth post on modeling the Coronavirus epidemic. I
recommend starting with the
first post and reading them in order.
So far, we’ve learned
about the SIR model and used
available data combined with the model to predict
the epidemic. We’re now going to detour into some additional
mathem...
Read more
This is my fourth post on modeling the Coronavirus epidemic. I
recommend starting with the
first post and reading them in order.
So far, we’ve learned
about the SIR model and used
available data combined with the model to predict
the epidemic. We’re now going to detour into some additional
mathem...
Read more
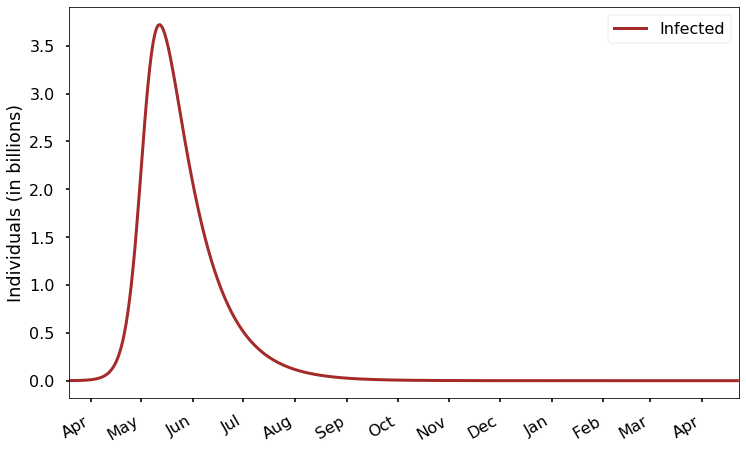
 Welcome to the third post in my series on modeling the Coronavirus
pandemic. In the first
and second
posts, we introduced the SIR model and used the available data to
estimate the model parameters, in the absence of any mitigation
(i.e. with no social distancing, quarantine, etc.). In this post w...
Read more
Welcome to the third post in my series on modeling the Coronavirus
pandemic. In the first
and second
posts, we introduced the SIR model and used the available data to
estimate the model parameters, in the absence of any mitigation
(i.e. with no social distancing, quarantine, etc.). In this post w...
Read more
19 Mar 2020
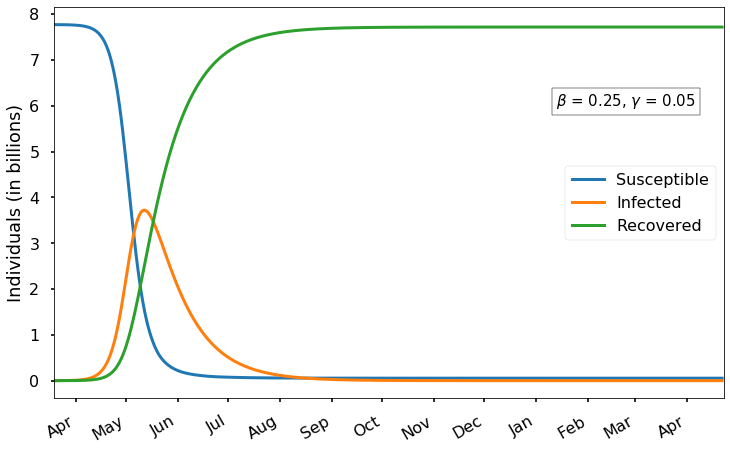
Welcome back! In the first post of this series, we learned about the SIR model, which consists of three differential equations describing the rate of change of susceptible (S), infected (I), and recovered (R) populations: \[\begin{align*} \frac{dS}{dt} & = -\beta I \frac{S}{N} \\ \frac{dI}{dt} & = \beta I \frac{S}{N}-\gamma I \\ \frac{dR}{dt} & = \gamma I \end{align*}\] As we discus...17 Mar 2020
This post is the first in a series in which we’ll use a simple but effective mathematical model to predict the ongoing Coronavirus outbreak. As I write, the number of officially confirmed global cases is just under 200,000 and many schools and workplaces all over the world have closed in order to slow its spread. If you’re like me, you’re wondering: Am I likely to catch this virus? How long wi...28 Mar 2016
Last Saturday, the ultramarathon scene finally, quietly, reached the Kingdom of Saudi Arabia. There was no fanfare and certainly no media coverage at this first organized footrace of more than marathon length. The course involved 50 kilometers of dirt, sand, gravel, and rocks in the hills northeast of Jeddah. An amazing group of more than 100 people showed up, including solo runners, relay team...29 May 2015
Welcome to step 7 in your training as a scientific Python code ninja: test automation. Tests are extremely useful, as long as you run them. Unfortunately, a common pattern in many projects is this: Tests are written and run by the person who wrote them. Others, and even the test-writer at a future date, forget to run the tests before committing new code. Or they decide not to run the tests bec...16 May 2015
Welcome to step 6 in your training as a scientific Python code ninja: issue tracking. Look at you! You’ve got your nice code with some docstrings and tests, and you’re humming along analyzing data, simulating widgets, and whatnot. In the middle of it all, you realize that your regression analysis function breaks if you feed it more than 57 data points. And wouldn’t it be nice if it made a plot ...15 May 2015
This is step 5 in your journey toward rock-solid scientific Python code: write tests. Tests? Ugh. I hear you. Writing tests is not an inherently fun process. However, the alternative is debugging, staring at code and thinking real hard about what could be wrong, and more debugging. I’ll take writing tests any day of the week. You may still not be convinced. Fortunately, I don’t have to convince...including examples! 14 May 2015
This is step 4 in your journey toward rock-solid scientific Python code. Steps 1-3 were language-agnostic, but now I’m going to assume you’re using Python. The Python language has a built-in feature for documenting functions, classes, and modules; it is the docstring. A docstring for a very simple function looks like this: def square(x): """ Takes a number x and returns x...13 May 2015
This is step 3 in your journey toward rock-solid scientific Python code. So your code is under version control and it’s floating up there in the cloud. In principle, someone else could use it. But how will they know it’s there? How will they know what it does, and how to install and use it? You need a README file. A README is minimalist documentation – just what is absolutely necessary for some...by putting your code in the cloud 12 May 2015
This is step 2 in your journey toward rock-solid scientific Python code. So you have your code under version control. Nice. But it’s sitting there on your computer. How do you share it with somebody else? And if they improve it, how do you incorporate their changes? Version control is your first tool in this regard, too. Your second tool is Github (or Bitbucket, Google Code, etc.). Github is a ...